Understanding the “error in glm.fit(x = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, : na/nan/inf in ‘y’]” Issue
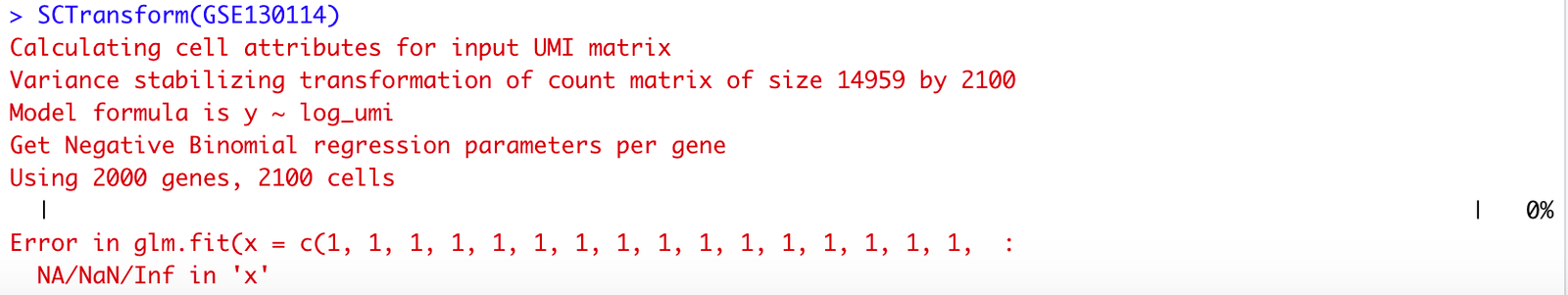
The error message “error in glm.fit(x = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, : na/nan/inf in ‘y’]” is one that frequently pops up when working with the Generalized Linear Model (GLM) function in R. This specific error indicates a problem with the input data, particularly in the response variable y, where values like NA, NaN, or Inf have caused the computation to fail.
In R, GLM is used to fit a generalized linear model, often for logistic regression, Poisson regression, or other statistical models. The model fitting process requires clean and valid data, but this error suggests that there is an issue with the data provided.
Causes of the Error
There are several potential causes behind the “error in glm.fit(x = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, : na/nan/inf in ‘y’]” message, most of which are data-related. Here are the primary reasons:
- Missing Values (
NA) in the Response Variable (y): If the response variable contains missing values (NA), the GLM function will fail to perform calculations. This is becauseNArepresents an unknown value, which prevents the model from making any meaningful predictions. - Non-numeric or Infinite Values (
NaNorInf) iny: Sometimes, the response variable may containNaN(Not a Number) orInf(infinite) values. These arise from undefined operations in the dataset, such as division by zero or other mathematical anomalies. GLM requires all values to be finite, so the presence of these invalid numbers will result in an error. - Issues with the Predictor Variables (
x): While the error message refers toy, the predictor variables (x) can also play a role. If the predictors contain constant values (e.g., all 1s) or similarly problematic data, it can lead to instability in the model fitting process, contributing to the same error. - Improper Data Types: Sometimes,
ymay be incorrectly specified as a factor or a character variable when the model expects a numeric or logical value. This mismatch can lead to errors during the fitting process.
How It Manifests
The “error in glm.fit(x = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, : na/nan/inf in ‘y’]” issue usually occurs during the execution of a model fitting command in R. Users typically experience this problem after they’ve run a GLM function such as:
glm(y ~ x1 + x2, data = mydata, family = binomial)
The error message appears abruptly, halting the code’s execution and preventing the user from obtaining any results. For those unfamiliar with the error, this can be frustrating, as it provides little information about the root cause.
Real-World Examples
Users have reported encountering this error in several scenarios. For instance, on forums like StackOverflow, one user noted that they received this error when trying to fit a logistic regression model. Upon investigation, they realized that their response variable had several NA values, which led to the breakdown.
Another user on an R-related forum discussed a similar issue when they attempted to build a Poisson regression model with infinite values in their dataset. These real-world examples highlight that the problem is common and largely arises from unclean or poorly prepared data.
Step-by-Step Guide to Resolving the Error
1. Check for Missing or Invalid Values in y
The first step in troubleshooting is to inspect the response variable (y) for missing (NA), undefined (NaN), or infinite (Inf) values. You can do this by running the following R command:
summary(y)
This will provide a summary of your variable, showing if any problematic values exist. If you discover NA values, you can choose to either remove them or impute them using a method like mean imputation. To remove NA values, use:
mydata <- na.omit(mydata)
For NaN or Inf values, you may need to either correct the underlying issue or exclude the rows that contain them:
mydata <- mydata[is.finite(mydata$y), ]
2. Ensure Proper Data Types
Verify that the response variable y is numeric or logical, depending on your model’s requirements. If it’s not, you can convert it as follows:
mydata$y <- as.numeric(mydata$y)
Ensuring the correct data type can often resolve the error when the problem lies in a mismatch between expected and actual types.
3. Inspect the Predictors (x)
If the error persists after cleaning the response variable, inspect the predictor variables to ensure they contain valid, non-constant data. Constant columns can lead to problems, as GLM expects some variance in the data. To check for constant columns, use:
sapply(mydata, function(x) length(unique(x)) > 1)
Remove any constant columns that do not contribute to the model:
mydata <- mydata[, sapply(mydata, function(x) length(unique(x)) > 1)]
4. Scale or Normalize the Data
In some cases, rescaling or normalizing the predictor variables can prevent instability in the model fitting process. This can be done using:
mydata <- scale(mydata)
This step ensures that all variables are on a similar scale, reducing the risk of overflow or underflow during calculations.
Tips to Prevent the Issue in the Future
- Clean Your Data Before Running Models: Regularly check your data for missing, undefined, or infinite values. Running a simple
summary()oris.finite()check can help you identify and address issues early on, preventing errors like “error in glm.fit(x = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, : na/nan/inf in ‘y’]” from arising. - Use Validation Techniques: Implement validation steps such as cross-validation or leave-one-out techniques to ensure your model is working correctly with different subsets of your data. This can help identify errors that only show up under certain conditions.
- Ensure Data Consistency: Keep your data types consistent, especially for the response variable
y. When preparing your data, always confirm thatyis numeric for GLM models unless specified otherwise. - Rescale and Normalize: If your dataset contains predictor variables with drastically different scales, rescale or normalize them before fitting your model. This practice not only prevents errors but also improves model performance.